SimFunctionSensitivity object
SimFunctionSensitivity object, subclass ofSimFunctionobject
Description
TheSimFunctionSensitivityobject is a subclass ofSimFunction object. It allows you to compute sensitivity.
Syntax
TheSimFunctionSensitivityobject shares all syntaxes of theSimFunctionobject. It has the following additional syntax.
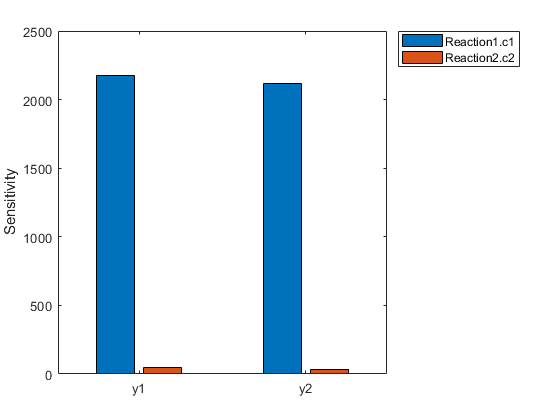
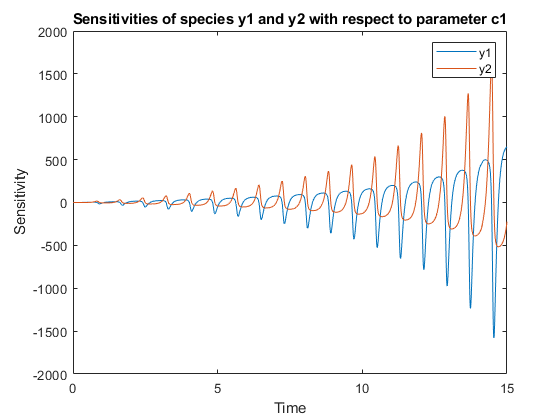
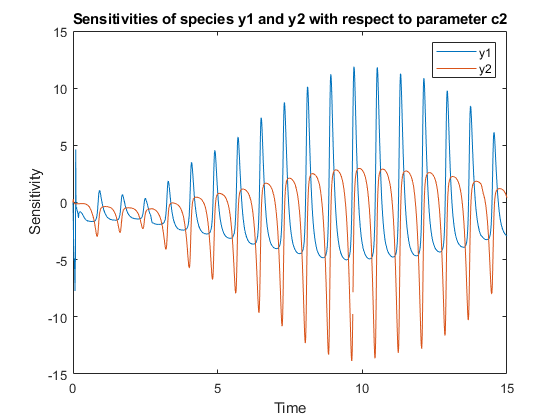
[T,Y,SensMatrix] = F(___)returnsT, a cell array of numeric vector,Y, a cell array of 2-D numeric matrices, andSensMatrix, a cell array of 3-D numeric matrix containing calculated sensitivities of model quantities.SensMatrixcontains a matrix of sizeTimePointsxOutputsxInputs.TimePointsis the total number of time points,Outputsis the total number of output factors, andInputsis the total number of input factors.
If you specify a single output argument, the object returns anSimDataobject or array ofSimDataobjects with sensitivity information.
Properties
TheSimFunctionSensitivityobject shares all properties of theSimFunction object. It has the following additional properties.
SensitivityOutputs |
This table contains information about model quantities (species or parameters) for which you want to compute the sensitivities. Sensitivity output factors are the numerators of time-dependent derivatives described inSensitivity Analysis in SimBiology. This property is read only. |
SensitivityInputs |
This table contains information about model quantities (species, compartments, or parameters) with respect to which you want to compute the sensitivities. Sensitivity input factors are the denominators of time-dependent derivatives described inSensitivity Analysis in SimBiology. This property is read only. |
SensitivityNormalization |
Character vector specifying the normalization method for calculated sensitivities. The following examples show how sensitivities of a speciesxwith respect to a parameterkare calculated for each normalization type.
|
![Figure contains an axes object. The axes object with title States versus Time contains 6 objects of type line. These objects represent y1, y2, d[y1]/d[Reaction1.c1], d[y2]/d[Reaction1.c1], d[y1]/d[Reaction2.c2], d[y2]/d[Reaction2.c2].](http://www.tatmou.com/help/examples/simbio/win64/CalculateSensitivitiesUsingSimFunctionSensitivityObjectExample_03.png)